Five weeks ago, I started working in the Bioconductor community, contributing to the BugSigDB Microbiome Study Curation project as an Outreachy intern. It has been such an experience, one filled with learning, growth, and exciting discoveries. Truly amazing and rewarding.
In this post, I will discuss the Human Microbiome, the BugSigDB project, what it is, how it works, and how anyone can get involved. I will also highlight the progress I have made so far as an intern, from reviewing and correcting curated studies to enhancing my skills in data science.
The Human Microbiome
The human microbiome is a complex collection of trillions of microorganisms, including bacteria, viruses, fungi, and other microbes, that live on and in our bodies. These microorganisms are mostly found in areas like our gut, skin, mouth, and respiratory and reproductive system. Though these microbes are as small as can be, they greatly impact our health. They help with essential functions like digestion, protecting us from harmful bacteria, and even influencing our immune system.
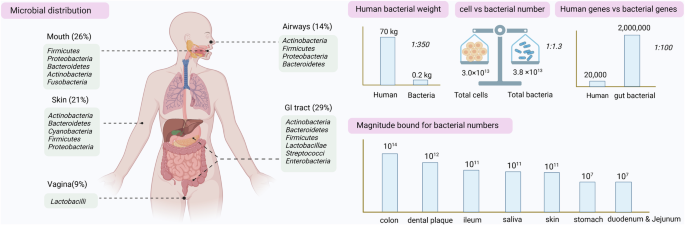
Recent research suggests that the microbiome might also play a role in mental health, potentially affecting conditions like depression and anxiety. For example, a study on gut dysbiosis found that it induces depression-like behavior through abnormal synapse pruning in microglia-mediated by complement C3 (Hao et al., Microbiome, 2024).
Fun fact💡*: I had the opportunity to curate this study in BugSigDB. You can find it* here.
The balance of these microbes is important for maintaining good health. When the microbiome is in balance, it works harmoniously with the body, promoting overall well-being. But when this balance is disrupted (dysbiosis), it can lead to health problems, such as autoimmune diseases, allergies, or gastrointestinal issues.
One of the main challenges in microbiome research is that the human microbiome is incredibly complex. Every individual has a unique microbial community, and factors like diet, lifestyle, medications, genetics, and environment all contribute to this variability. As more and more studies are published, the amount of data grows rapidly, making it harder for researchers to make sense of all the findings. This is where tools like BugSigDB come in, helping researchers organize, analyze, and extract meaningful data from the extensive amount of microbial data available.
BugSigDB: Simplifying Microbiome Research

BugSigDB is a comprehensive community-editable database designed to help make sense of the enormous amounts of data related to the human microbiome. As researchers continue to uncover the complex relationships between microbiota and human health, BugSigDB stores these microbial signatures associated with various health conditions. The microbial signatures are essentially patterns in the composition of the microbiome that are linked to specific diseases or health outcomes.
What makes BugSigDB particularly valuable is its ability to simplify the analysis of microbial data. Instead of manually searching through countless studies and publications, researchers can use BugSigDB to find microbial signatures linked to certain diseases quickly. This saves time and helps to effectively organize the research process. BugSigDB also allows for the identification of trends across multiple studies, making it easier to spot consistent patterns in microbial composition that may contribute to health conditions.
BugSigDB includes but is not limited to studies covering:
Health Conditions: Microbial patterns in diseases like diabetes, cancers, neurological disorders etc.
Drugs and Treatments: How antibiotics, therapies and treatments in general affect the microbiome.
Experimental Studies: Studies where samples have been taken from human body sites or animal models.
Key Features of BugSigDB
Designed to make microbiome research more efficient and accessible, here are some of the key features of BugSigDB:
1. Open-Source and Community-Driven

Screenshot from BugSigDB REPO on GitHub
BugSigDB is open-source, meaning anyone can contribute, access or review existing data. This collaborative approach ensures the database stays accurate and grows with the latest research.
2. Easy-to-Use

The database is designed to be user-friendly. Users can search for studies based on health conditions, location, signatures, host species, body sites, or even by study design, making it simple for both beginners and experts to use.
3. Quality
Every entry is reviewed to ensure reliability. This process helps maintain high-quality, trustworthy information for researchers using BugSigDB.

Screenshot from BugSigDB REPO on GitHub
Fun fact 💡*:* Issue #74 was the first issue I requested to work on during my initial contribution to BugSigDB for the Dec-March 2023 Outreachy cohort. How time flies indeed!
4. Multiple Studies
In BugSigDB, you will find a wide range of studies, from health outcomes to drug effects, time series, case-control, studies on animal models, and various diseases making it a comprehensive resource for anyone studying the microbiome.
This simplicity and versatility makes BugSigDB an essential tool in microbiome research. For more details, the article BugSigDB captures patterns of differential abundance across a broad range of host-associated microbial signatures (Geistlinger et al., Nat Biotechnol., 2024.) offers an in-depth look at the project.
Contributing to BugSigDB
BugSigDB organizes microbial signatures from studies into a searchable format. Using a system similar to Wikipedia, BugSigDB makes it easy for people to edit and update data. This keeps the database fresh and relevant. Here’s an overview of how it works and ways you can contribute:
The Curation and Review Process
Find a Paper of Interest that has not been added to BugSigDB yet. Look for curatable information, such as:
Study design
Contrast groups
Statistical tests
Alpha diversity measures
Significant differentially abundant microbial signatures
Such studies can be found in journals such as Nature Biotechnology, Microbiome, American Heart Association, Science Direct, Wiley, PubMed, American Society Microbiology, etc.
Alternatively, you can choose a paper from the BugSigDB curation GitHub REPO (these papers have already been vetted and found to be curatable with relevant data on the human microbiome).
Once you’ve selected a study, proceed with curation by adding:
Study information
Experiments
Microbial signatures
If you are unsure about any step, refer to the tutorial videos for guidance. And don’t forget, help is always available! If you’re stuck, feel free to reach out on Slack for support from the community.
Submit for Review: After curating the study, submit it for review. A reviewer will verify the accuracy of your entries and provide feedback on areas that may need correction or improvement.
Correct Based on Feedback: Address any feedback provided by the reviewer to ensure the data meets BugSigDB’s standards. This step is important for maintaining the quality and reliability of BugSigDB.

- Final Approval: Once all feedback has been addressed, the study will be marked as reviewed, and the issue closed. The study is now officially part of the BugSigDB database!
BugSigDB in Action
To see BugSigDB’s impact, let’s look at an example.
A researcher studying obesity notices that certain signature like Akkermansia muciniphila for example, appears to be more abundant in individuals with higher body mass index (BMI). However, confirming this finding across different populations and studies would be challenging and time-consuming.
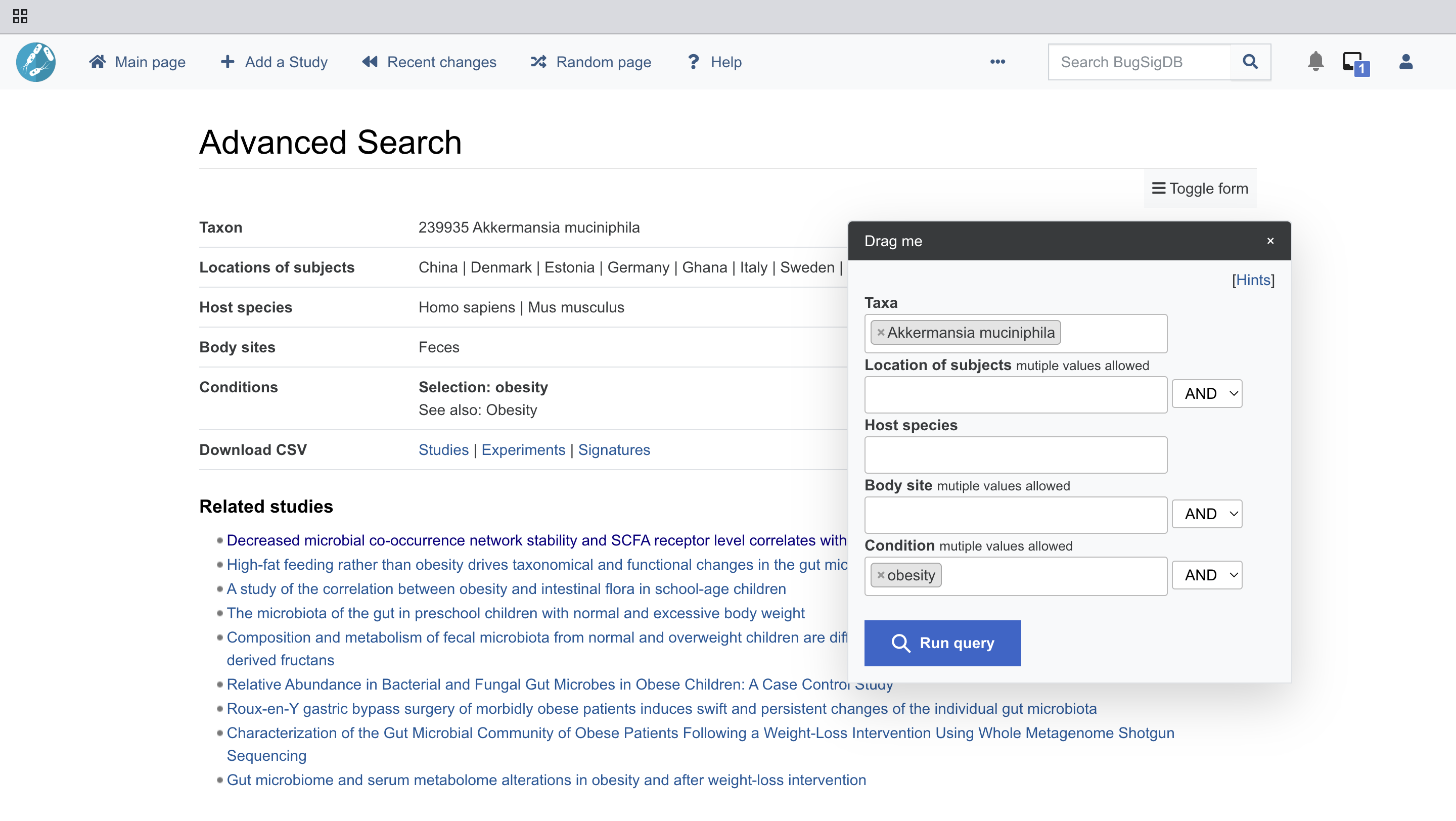
With BugSigDB, the researcher can quickly access a collection of studies that link Akkermansia muciniphila to obesity. The database not only confirms their initial observation but also uncovers other microbial patterns related to weight gain and metabolic health. This accelerates the research process and strengthens the foundation for further investigations into the role of the human microbiome in obesity.
My Journey as an Intern
Over the course of my internship, I have had the privilege of working on different research papers relating to the human microbiome, this experience has deepened my understanding of the human microbiome and has also allowed me to grow my skills in data science and research. From reviewing studies to expanding my programming knowledge, every task has been an opportunity to learn and contribute.
A closer look at the progress I’ve made so far:
Reviews and Corrections
As a contributor to the BugSigDB project, I have completed the curation, reviews, and corrections of over 80 studies in BugSigDB. This has been a great opportunity to contribute to the quality and reliability of BugSigDB while honing my attention to detail.

Screenshot from BugSigDB REPO on GitHub
Expanding Data in BugSigDB
I have also focused on increasing data for underrepresented conditions. Specifically, I have located and read published research papers related to hypoglycemia (Low blood glucose) and hypotension (low blood pressure), identifying them for curation in BugSigDB. This helps to make the database more comprehensive and valuable for anyone exploring these conditions.
Feel free to suggest or add papers to the current list I have curated, or reach out if there is any condition of interest you’d like to see included in the BugSigDB database. I’d be happy to share ideas!
Advancing My R Programming Skills
To take on more advanced tasks, I have been leveling up my R programming skills. This is preparing me to work with data analysis and handling tasks within BugSigDB, which is in perfect alignment with my growing interest in data science. Exploring R has been exciting, as it is such a powerful tool for uncovering patterns and gaining understanding into large datasets.
Pursuing Extracurricular Learning
To support my progress, I have also commenced with courses on linear algebra and statistics. These have been really helpful for understanding and interpreting data in a more structured way. Both help with visualizing and transforming data for better interpretation of trends and patterns. Both of these are essential for making sense of microbiome data and studies.
Want to Contribute to BugSigDB?
One of the best parts of BugSigDB is that anyone can get involved! Doesn’t matter if you are a researcher, student, or microbiome enthusiast, there are many ways you can get involved and make an impact:
1. Join the Community
Start by becoming part of the BugSigDB community! Connect with like-minded contributors through the Slack channel.
2. Explore the Database
Familiarize yourself with BugSigDB by exploring the website and the curated studies. Understanding how studies are curated will help you become an effective contributor. Resources like the [Curation Policy](curation Policy - BugSigDB) and videos are great starting points.
3. Request an Account
To contribute, you will need an approved account. Submit a request to gain access and begin exploring the tools and resources available for curation and analysis.
4. Curate New Studies
Step into microbiome research by curating new studies. This involves adding study details, experiments, and microbial signatures to the database, ensuring they meet BugSigDB’s curation policy.
5. Analyze and Compare Studies
Analyze and compare curated studies to extract valuable data, identify patterns, and support your research using the bugsigdbr R/Bioconductor package.
6. Feature Requests and Enhancements
BugSigDB is under active development and we are very open to feature requests and enhancements to improve its usefulness for researchers and users. You can post feature requests on the project GitHub or contact us via the Google Group.

Source: GIPHY
Contributing to BugSigDB is an exciting opportunity to support microbiome research while developing valuable skills and knowledge of the human microbiome. Whether you're curating, reviewing, or analyzing, every effort helps advance the field and improve the BugSigDB’s usability for the global research community. Again, every contribution counts!